Warum RechnungFertig?
Weil viele Einzelunternehmer und Selbständige nur ein einfaches Rechnungsprogramm benötigen
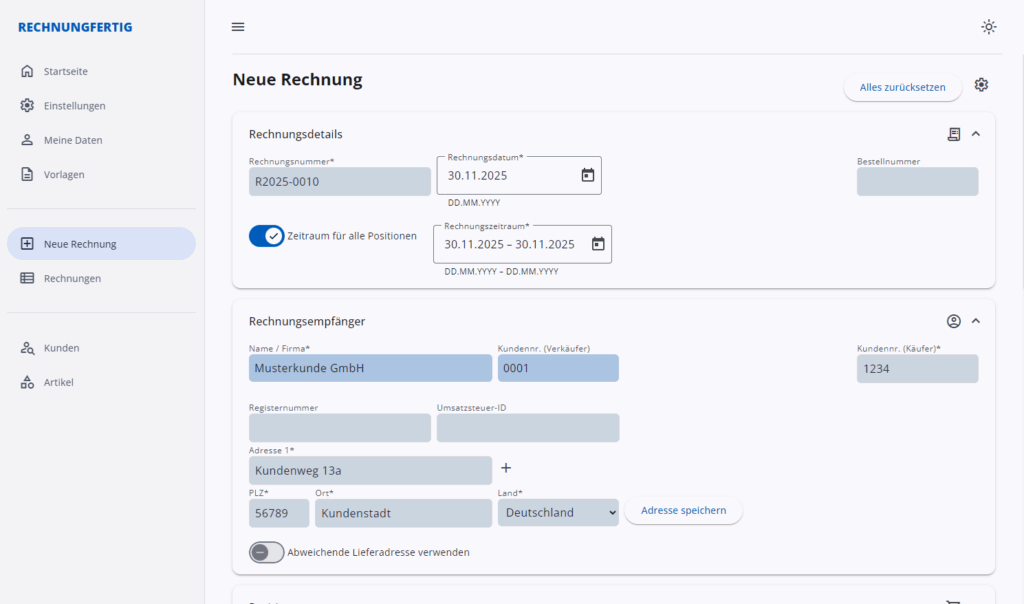

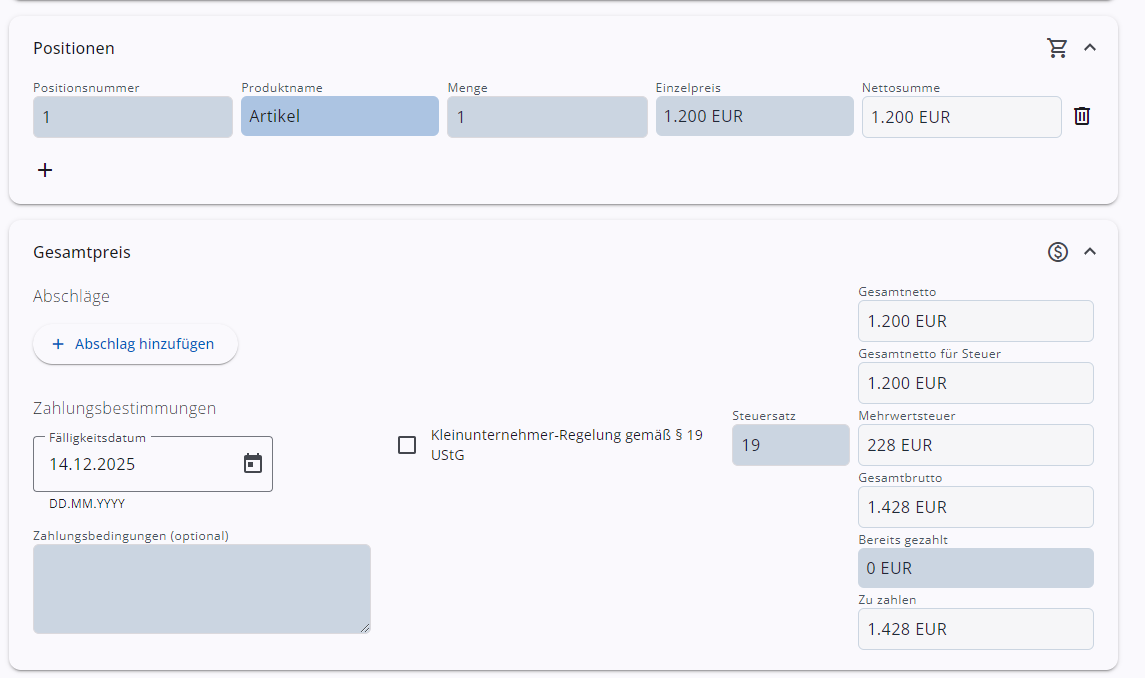
Rechnungen erstellen
Erstelle Rechnungen und verschicke sie als ZUGFeRD-Datei (PDF-Format mit angehängter Factur-X XML-Datei). mehr…
Rechnungslayouts
Templates für Rechnungen können in Word erstellt und für die Erzeugung der PDF-Rechnung benutzt werden. mehr…
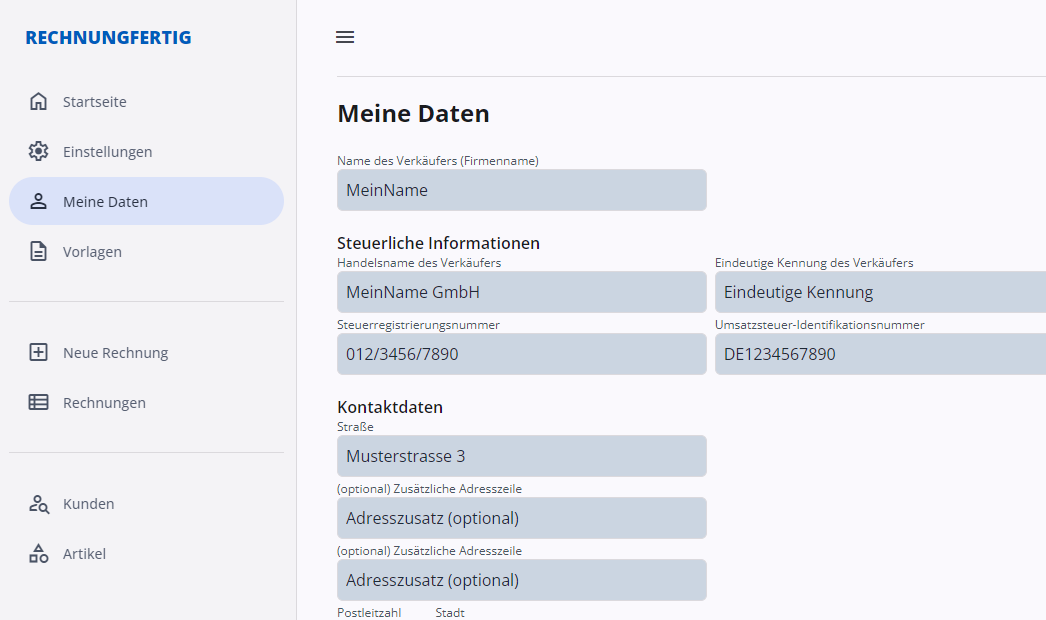
Keine Cloud
Alle Daten werden lokal gespeichert und können auch mit anderen Programmen geöffnet werden. mehr…

Kein Abo
Kein Abo, keine Werbung. Sie kaufen einmal die Software und können sie danach solange benutzen, wie Sie wollen. mehr…

An wen richtet sich RechnungFertig?
RechnungFertig richtet sich vor allem an Selbständige und Einzelunternehmer, die bislang gut damit zurecht kamen, ihre Rechnungen in Word zu erstellen und jetzt einfach nur ein günstiges Rechnungsprogramm suchen, um die technischen Anforderungen der E-Rechnung zu erfüllen. Zum Beispiel:

Berater

Coaches

Nebenberufstätige

Produktanbieter
E-Rechnungspflicht & ZUGFeRD – einfach gesetzeskonform bleiben
Mit der schrittweisen E-Rechnungspflicht ab 2025 müssen Selbstständige und Unternehmen Rechnungen zunehmend in strukturierten elektronischen Formaten wie ZUGFeRD oder XRechnung erstellen – insbesondere im B2B-Umfeld und bei öffentlichen Auftraggebern. RechnungFertig übernimmt diese Anforderungen automatisch im Hintergrund, sodass Sie weiterhin einfach arbeiten, aber rechtssicher vorbereitet sind. Offizielle Informationen zur E-Rechnung stellt das Bundesministerium der Finanzen bereit.

Aktuelle Blog-Artikel
-
ZUGFeRD und Co: 10 Begriffe zum Thema E-Rechnung
Wer sich als Selbständiger oder Kleinunternehmer gerade mit der E-Rechnungspflicht und den neuen Abläufen beschäftigt, stolpert schnell über einen Begriffsdschungel: ZUGFeRD, Factur-X, XRechnung, EN 16931 sowie Profile wie BASIC und BASIC WL. Aber was bedeuten diese Begriffe? Hier folgt eine kleine Einordnung, bei der das Schaubild hoffentlich etwas Licht ins Dunkel bringen kann. ZUGFeRD und…
-
Update 1.5.0: Rechnungsarten und XRechnung
Version 1.5.0 von RechnungFertig ist draussen mit Updates, die für Selbständige, Kleinunternehmer und Nebenberufler einiges verbessern: Rechnungstypen Ein Kernpunkt sind Rechnungen mit verschiedenen Rechnungstypen (Business Term 3). Damit lassen sich unterschiedliche Geschäftsfälle klarer abbilden, ohne Workarounds oder manuelle Umbenennungen im Dokument. Durch ein Popup, das über das Fragezeichensymbol aufrufbar ist, gibt es zudem weitere Erklärungen…
-
E-Rechnung für Kleinunternehmer
Seit 01.01.2025 ist „E-Rechnung“ in Deutschland nicht mehr gleichbedeutend mit „PDF per E-Mail“, sondern meint grundsätzlich strukturierte Rechnungsdaten nach EN 16931 – z. B. XRechnung oder ZUGFeRD (ab 2.0.1), je nach Profil. Für dich als Kleinunternehmer nach § 19 UStG ist das doppelt wichtig: Du musst E-Rechnungen empfangen können (organisatorisch reicht oft ein E-Mail-Postfach, die weitere…
Neues zu RechnungFertig
RechnungFertig wird regelmässig aktualisiert. Da die Software selbst nicht auf das Internet zugreift, können Sie sich hier für den Newsletter eintragen, um informiert zu werden, wenn es ein neues Update für RechnungFertig gibt.