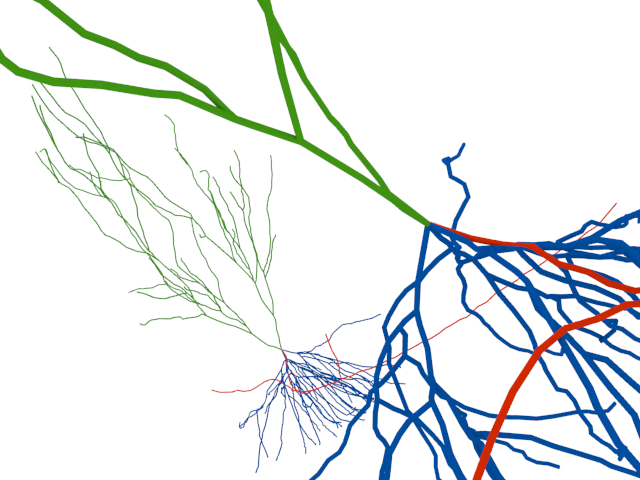
 The swc-importer for neural reconstructions is also now on Github. I used this opportunity to embed the key function into an importer class for Blender.
The swc-importer for neural reconstructions is also now on Github. I used this opportunity to embed the key function into an importer class for Blender.
Martin Pyka
Neuroscience, Data Analysis, Art, Stuff

 The swc-importer for neural reconstructions is also now on Github. I used this opportunity to embed the key function into an importer class for Blender.
The swc-importer for neural reconstructions is also now on Github. I used this opportunity to embed the key function into an importer class for Blender.
Dear Dr. Pyka,
I have recently been attempting to import some neuronal swc files into Blender 2.78c, using your SWC2Blender addon (https://github.com/MartinPyka/SWC2Blender)
I think I have installed it in Blender correctly because it works, sort of (!). The morphology of the neurons appear to be correct (compared to how they look in a basic swc editing program, Neutube), but for some reason the segment radii are not read correctly.
I have tried to import both the example neurons in the folder. In both cases the thick cell body segments are almost invisible, and thicker dendrites are clearly thinner than they should be.
I have also tried various older versions of Blender including 2.74 (for which the plugin was written?) with no difference. Do you have any ideas what might be going wrong? I hesitate to suggest that there is something wrong with the addon itself –
it is way more likely that I am doing something stupid out of sheer ignorance! But could you check to see if the addon still works properly for Blender 2.78c?
Thanks a lot for your time, and for all your work in making this tool!
sorry for my late reply but I am not checking the comments in my blog very often.
I am not working in this area any more and have no time to check, but the two crucial lines in the code are these one:
https://github.com/MartinPyka/SWC2Blender/blob/master/io_mesh_swc/operator_swc_import.py#L7
https://github.com/MartinPyka/SWC2Blender/blob/master/io_mesh_swc/operator_swc_import.py#L65
you might want to check for your own, whether there is something wrong in the code. I remember, that I essentially only multiply what is given in the swc file by a constant factor.
Best,
Martin
Hej Martin, your code was really useful for visualising neurons, however there is a bug where the radie increases with increased compartment number. The code should use value[4] instead of value[5] since the first column is removed when inserted into the dictionary. Also the soma is missing from the visualisation.
I have an updated version of the import script, can email it to you if you want to update your github.
Cheers,
Johannes